Chemicals Assessment and Research Center
- Home
- Chemicals Assessment and Research Center
- CARCINO screen®
CARCINO screen®
CARCINOscreen®
- What is CARCINOscreen®?
- Highlights of CARCINOscreen®
- Service Menu for CARCINOscreen®
- Option Services
- Deliverables/Delivery date
- Prices
- Ordering
- History of CARCINOscreen® development
- Simpler alternative to CARCINOscreen® based on quantitative PCR (qPCR)
What is CARCINOscreen®?
Short-term carcinogenicity prediction system; CARCINOscreen® offers the predictive information for the carcinogenic potential of chemcials by the gene expression profiles in the liver of rats obtained in short-term animal experiments (14days or 28 days) (Fig. 1) (Matsumoto et al., 2014).
-
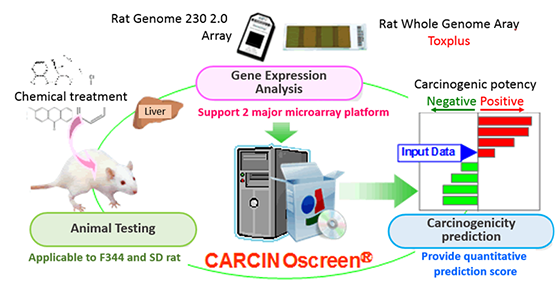
- Fig.1 Overview of CARCINOscreen®
yApplicable Conditionsz
| Species/Strain/Sex | Crlj:CD (SD) rat or F344 rat, male |
|---|---|
| Organ | Liver |
| Number of Animals for Gene Expression Analysis |
3 animals/group |
| Dosage | Concurrent vehicle control + Chemical treatment groups (more than 2 groups) |
| Route of Administration | gavage |
| Duration of Treatment | 14 days or 28 days |
| Microarray Platform | EWhole Rat Genome Toxplus (Agilent, Custom Array) ERat Genome 230 2.0 Array (GeneChip®; Affymetrix) |
Highlights of CARCINOscreen®
- Short-term, High-accuracy and Low-cost as carcinogenicity screening
Accuracy (concordance) is more than 90%.
Quantitative prediction results can be obtained as shown in Fig. 2. - Applicable to both Ames-positive carcinogens and Ames-negative carcinogens (Fig.2)
-
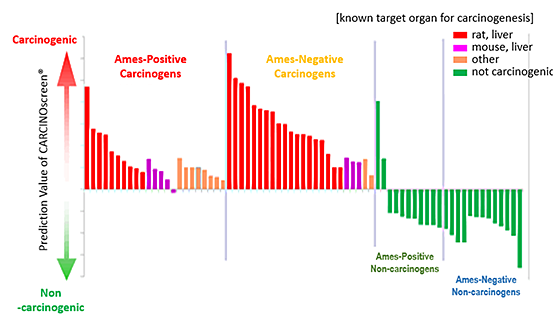
- Fig.2 Prediction results by CARCINOscreen® (73 chemicals)
- Various services
Depending on your experimental design, we can offer various types of services (strain, treatment duration, microarray platform). - Allow you a comprehensive gene expression analysis at the same time
Service Menu for CARCINOscreen®
Table 1 Service menu for Short-term carcinogenicity screening iCARCINOscreen®j
| Menu# | Strain | Treatment duration |
Microarray platform |
Notes |
|---|---|---|---|---|
| Menu-(1) | F344 rat | 28 days | Toxplus array*2 (Agilent) |
- |
| Menu-(2) | SD rat*1 | 28 days | Applicable to generally used strain for animal toxicity testing | |
| Menu-(3) | F344 rat SD rat*1 |
14 days | GeneChip® (Affymetrix) |
|
*1FCrlj:CD (SD) rat
*2FWhole Rat Genome Toxplus (Agilent, custom array)
Menu-(1) Prediction using Toxplus array, applying 28-day treatment sample in F344 rat
This menu is applicable to the liver sample obtained from F344 rat. Whole Rat Genome Array Toxplus (herein after Toxplus array) is used for gene expression analysis. Toxplus array is the specifically designed microarray, carrying prediction genes customized for carcinogenicity prediction by CARCINOscreen® and whole genes (approximately 40,000 genes) on “Whole Rat Genome array (Agilent)”. This array offer you not only the predictive result for carcinogenicity, but also a comprehensive gene expression analysis data, at the same time.
Menu-(2) Prediction using Toxplus array, applying 28-day treatment sample in SD rat
This menu is achieved by expanding the application of [Menu (1)] to SD rat that is generally used for animal toxicity testing. Toxplus array is used for the gene expression analysis. To extend [Menu (1)] application, the system was validated using 13 chemicals that had been tested in F344 rat. The results revealed that all liver carcinogens were detected (9/9 chemicals), and that 8 of them were Ames-negative liver carcinogens (Table 2). Among 13 chemicals, 11 chemicals were correctly predicted and this system is judged to be also applicable to the liver from SD rat.
Table 2@Prediction results of 13 chemicals in SD rat
| Chemical Name | Known information | Prediction results by CARCINOscreen® | |||
|---|---|---|---|---|---|
| Carc.* | Rat | Ames | F344 rat | SD rat | |
| Liver | |||||
| Carc. | |||||
| 2,4-Diaminotoluene | + | + | + | positive | positive |
| Methyl carbamate | + | + | | | positive | positive |
| Thioacetamide | + | + | | | positive | positive |
| Urethane | + | + | | | positive | positive |
| D,L-Ethionine | + | + | | | positive | positive |
| Chlorendic acid | + | + | | | positive | positive |
| Clofibrate | + | + | | | positive | positive |
| Di(2-ethylhexyl)phthalate (DEHP) |
+ | + | | | positive | positive |
| 3-(p-Chlorophenyl)- 1,1-dimethylurea |
+ | + | | | positive | positive |
| Di(2-ethylhexyl)adipate (DEHA) |
+ | | | | | positive | negative |
| 2,6-Diaminotoluene | | | | | + | negative | negative |
| 2-Chloromethylpyridine HCl | | | | | + | negative | negative |
| Iodoform | | | | | + | negative | positive |
*:Carcinogneicity
Menu-(3) Prediction using GeneChip®, applying 14-day treatment sample
This menu is applicable to the test sample obtained from shorter-treatment, i.e., 14-day. GeneChip® is applied for gene expression analysis. This menu allow you the use of liver samples from a preliminary test (e.g., dose setting test) of toxicity testing. If you have already measured the gene expression data by GeneChip®, the data can also be applied to our system.
Option Services
- From animal testing and microarray analysis to carcinogenicity prediction by CARCINOscreen®.
- From microarray experiment to prediction, if you already have liver samples.
- Carcinogenicity prediction only, if you already have gene expression data (Toxplus array or GeneChip®)
-
- Options for placing your order
yFlow for your orderz
| 1) Present our analysis plan | We will make plan according to your purpose and experimental system. |
| 2) Prepare cost estimation | We will present the cost estimation according to the plan. |
| 3) Receipt your sample or data | Please send us your sample (chemical, organ sample (stored in RNA later), or total RNA) or data file depending on your plan. |
| 4) Animal testing/ microarray experiment/ carcinogenicity prediction |
Experiments are conducted according to your plan. |
| 5) Deriver the outcomes | The report for each experiment will be prepared and delivered with all data file obtained. |
Deliverables/Delivery Date
In addition to the report (printed matter), we will deliver the following data as an electronic file. The contents of the report includes (summary), purpose, method, results (discussion).
| Deliverables | (1) Test report (PDF file) |
|---|---|
| Delivery Date | Please contact us. |
Prices
Please contact us.
Ordering
- Please provide us your information (purpose of the analysis, experimental conditions etc.) as much as possible using this form. Specific analysis conditions will be determined after consultation with you.
- We will apply the contracts to provide our service.
History of CARCINOscreen® Development
We, CERI, had developed a carcinogenicity prediction method based on gene expression analysis obtained from microarray in NEDO Toxicogenomics Project in 2001-2006 (lead by Professor T. Shirai (Nagoya City Univercity). The resultant microarray data in the project were registered in GEO (Gene Expression Omnibus) database in NCBI (National Center for Biotechnology Information) (available 3-day treatment dataA14-day treatment dataA28-day treatment data). Based on the results from this NEDO project and our additional research, CERI put them into a practical use as a carcinogenicity prediction screening tool, named as CARCINOscreen®, and launched its service in 2010.
Simpler alternative to CARCINOscreen® based on quantitative PCR (qPCR)
For the purpose of more simple prediction, we minimize the prediction genes used in CARCINOscreen® and developed the qPCR based prediction method (Saito et al., 2016) in Tox-Omics project in 2012-2016. This method achieves qualitative carcinogenicity prediction with low cost than CARCINOscreen®. The experimental protocol can be found from here.
![CERI ๊สเc@l ปwจฟ]ฟค@\](../../img/ceri_logo.png)